SNPs have recently become markers of choice because of their abundance in the genome. High throughput NGS technologies also prolifically aided in deciphering millions of SNPs genome wide. This database is an attempt to club this information and classify the SNPs based on their distribution in the genome.
This public domain database archives over a 1.9 million variations spanning across the genome, deciphered from 29 whole genome re-sequenced (WGRS) and 61 restriction site-associated DNA sequenced (RAD) lines of chickpea. These variations (substitutions, small length insertions and deletions) were identified from upstream and downstream regions of genes, intronic, exonic and intergenic regions of genome. The database offers one-stop destination to the chickpea research community to exploit these sequence variations within genomes for a better understanding of molecular genetics, gene mapping, defining population structure and other functional studies. The database is implemented with webBlast and JBrowse which makes it even more congenial and streamlined for its users.
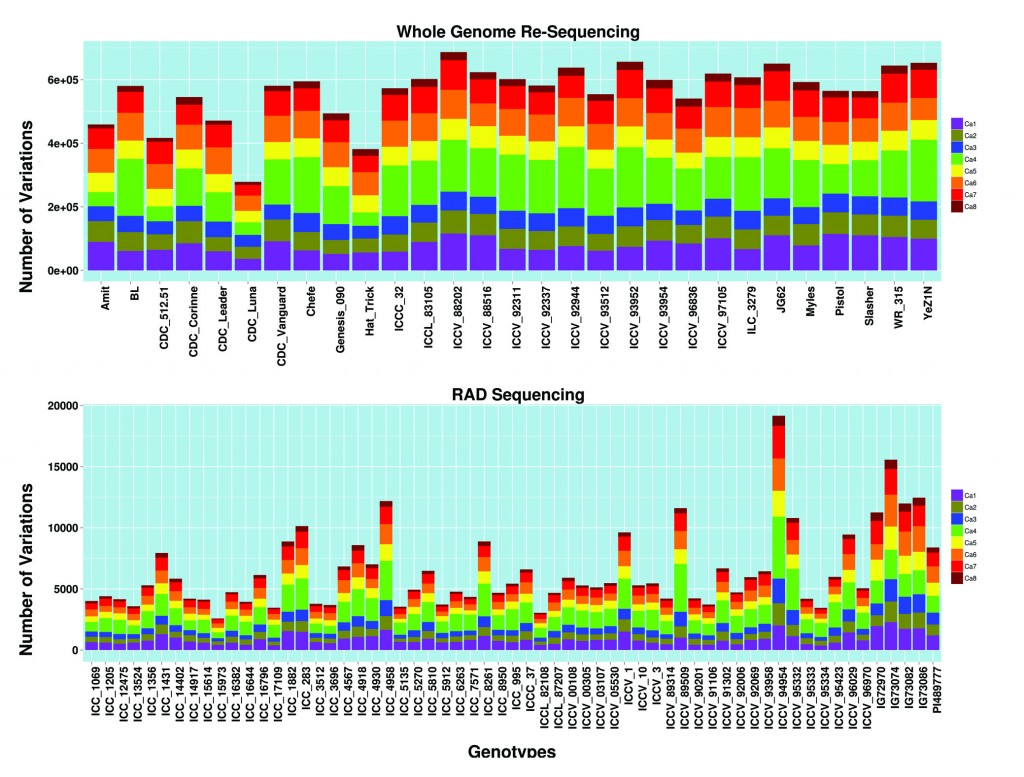
Doddamani D., Khan A.W., Katta M.A.V.S.K., Agarwal G., Thudi M., Ruperao P., Edwards D. and Varshney R.K. (2015) CicArVarDB: SNP and InDel database for advancing genetics research and breeding applications in chickpea. Database. 01-07. ISSN 1758-0463
